IS1222: analysis and distribution of a new insertion sequence in Enterobacter agglomerans 339
(Insertion sequences IS407, IS476 and ISR1; IS3 family; translational frameshifting; pseudoknot; Rahnella aquatilis)
Hans-Dieter Steibl/a and Frank-Michael Lewecke/b
a Lehrstuhl für Genetik, Universität Bayreuth, D-95440 Bayreuth, Germany, Tel. (49-921)552702, Fax (49-921)552535; and b Medizinische Klinik IV, Universität Erlangen-Nürnberg, Krankenhausstraße 12, D-91054 Erlangen, Germany
Correspondence to: F.-M. Lewecke, Medizinische Klinik IV, Universität Erlangen-Nürnberg, Krankenhausstraße 12, D-91054 Erlangen (Germany). Tel. (49-9131) 859002; Fax (49-9131) 859209
Introduction
Experimental and Discussion
Acknowledgements
References
Figures and Tables
Abbreviations: aa, amino acid(s); bp, base pair(s); E., Enterobacter; GCG, Genetics Computer Group (Madison, WI, USA); IR, inverted repeat; IS, insertion sequence(s) (Fiandt et al., 1972); kb, kilobase(s) or 1000 bp; nif, nitrogen-fixation gene; nt, nucleotide(s); ORF (Orf, orf), open reading frame; P., Pseudomonas; RBS, ribosome-binding site(s).
With a length of 1221 bp and 44-bp inverted repeats with ten mismatches, IS1222 was identified as an endogenous insertion sequence in Enterobacter agglomerans 339. In this host strain, four copies were located, three on the nif plasmid pEA9 and one at the chromosome. Sequence analysis showed two consecutive open reading frames, orfA and orfB, encoding putative polypeptides of 87 and 276 amino acids. Inbetween both reading frames, a potential frameshift window of the homonucleotide type was postulated, followed by a pseudoknot structure and a ribosome-binding site. Based on significant homology at the sequence level and similarity of the features discussed, IS1222 was placed among the group of IS3 elements with IS407, IS476 and ISR1 being the most closely related IS. Hybridization experiments suggest that the distribution of IS1222 is limited to a group of related bacterial strains among Enterobacteriaceae.
Different nitrogen-fixing Enterobacter agglomerans (E. agglomerans) strains isolated from the rhizosphere of barley and wheat in the vicinity of Bayreuth (Bavaria, Germany) were characterized in our workgroup. Detailed analysis revealed a specific, clustered organization of nitrogen-fixation (nif) genes located on large plasmids (Kleeberger et al., 1983; Singh et al., 1983). In further studies the 200-kb nif plasmid pEA9 from E. agglomerans 339 was shown to be self-transmissible (Klingmüller et al., 1989), and in order to characterize nif-flanking genes, the 3'-flanking region of the pEA9 nif cluster was sequenced. Database searching in EMBL/GenBank with the obtained nt sequences of two ORFs showed significant homology to orfA and orfB of insertion sequences (IS) of the IS3 family (Steibl, 1994).
Apart from prominent features shared by all IS elements, such as terminal inverted repeats (IR) or the ability to transpose to many sites on bacterial plasmids and chromosomes, this particular group of IS share a common structure with a small ORF in phase 0, followed by a larger ORF in phase -1. With a similar size of 1.2- to 1.5-kb IS3 elements are most widespread among different taxonomic genera of host organisms. Conventionally the orientation of the IS is defined by the a and b termini and orfA, orfB respectively (OrfA, OrfB for the gene products) as the coding reading frames (Polard et al., 1991; Chandler and Fayet, 1993).
Interest in the IS3 family has been increasing since similarities to retroviral features became apparent. Both groups terminate in 5'-TG-3', the process of element-specific gene expression involves a -1 translational frameshift, and both retroviral insertion and the transposition of IS share similar mechanisms (Fayet et al., 1990; Khan et al., 1990).
Here we report the presence of a novel IS3 element in Enterobacter (assignment of IS1222 by the Plasmid Reference Center, Stanford, CA, USA), displaying a possible motif for translational frameshifting of the homonucleotide type (Chandler and Fayet, 1993; Sekine et al., 1994).
(a) Construction of plasmids containing IS1222
From the BamHI-cosmid library of the indigenous 200-kb plasmid pEA9 the cosmid pHD54 contains the complete nif cluster including both flanking regions (Steibl et al., 1993). For a detailed analysis of the 3'-flanking region, restriction fragments from pHD54 were subcloned (Fig. 1).
(b) Relevant features of IS1222
The nt sequence of IS1222 is shown in Fig. 2, including both flanking regions. The IS element is 1221 bp in length, has 44-bp imperfect terminal IR with ten mismatches and is not flanked by target site duplications (direct repeats), possibly caused by secondary mutations like the replacement of GA in the putative 5'-149AGAG153-3' repeat. For the entire IS a G+C content of 56 % was determined, which makes it a distinct structure in between the flanking sequences with an average of 45% G+C. Sequence analysis revealed two major ORFs on both strands, but as shown for related IS, only those ORFs, coding from a towards b terminus (orfA/orfB and insA/insB, respectively), were of significance (Polard et al., 1991; Vögele et al., 1991). Accordingly, features of orfA and orfB will be discussed only. OrfA, predicting a protein of 87 aa, extends from nt 224 up to nt 487. The orfB coding in phase -1 relative to orfA starts at nt 514 and stops at nt 1344. Translation of orfB results in a 276-aa protein. The search for s70-like promoter structures revealed two possible signals for transcription initiation, named P1 and P2. P1 at nt 165 is followed by a RBS and situated within the left IR. Downstream from P2, located on the opposite strand at nt 1066, no signal structures for translation start in combination with a reading frame were found, although transcription of genes outside the IS might be activated. Starting at nt 193 an IR was discovered between P1 and the RBS ahead of orfA. Only in case of transcription from upstream into the IS the mRNA could form a stable hairpin with a stem length of 10 nt and a loop of 7 nt (DG = -10.0 kcal/mol, calculated using the GCG-program FOLD; Zuker and Stiegler, 1981). Lying within the stem structure, the RBS could then be obscured preventing translation of orfA (Fig. 2). The underlying mechanism would be a means of controlling transposition as postulated for IS150 (Schwartz et al., 1988).
(c) Characterization of OrfA and OrfB
OrfA with a size of 87 aa displays a highly positive charge with a pI=10.42 as calculated by the GCG program ISOELECTRIC (Devereux et al., 1984). Apart from this significant characteristic for DNA-binding proteins, a potential ahelix-turn-ahelix motif was located extending from aa 23 to 42. Using the frequency and weight matrix table for highly conserved aa described by Dodd and Egan (1987) a score value of 1230 was determined, representing a high probability for the actual existence of a DNA-binding motif. Similar score values were calculated for related IS3 elements by Prère et al. (1990).
For the large OrfB with 276 aa an equally high charge (pI=10.33) was calculated. Further sequence analysis on aa level revealed a D(35)E motif between aa 172 and 209. This region, highly conserved for retroviruses and so far most IS3 elements and encompassing about 50 residues, contains a central feature with a conserved D followed by 35 aa and then a conserved E. According to Khan et al. (1990) the potential for DNA binding, together with MgATP and phosphate transfer associated with D and E residues in other proteins, suggests that the D(35)E sequences in integrase and OrfB take part in binding, cutting and transfer of DNA strands (Fayet et al., 1990; Khan et al., 1990).
(d) Comparison with IS3 elements
For comparative analysis of IS1222 the following well studied IS elements were selected from the group of IS3 elements characterized so far: IS407, IS476, ISR1, IS150, IS861, IS3, IS911 and IS3411 (references and emendation of nt sequences in Prère et al., 1990). IS were compared in respect to their terminal IR (Table I) and conservation of OrfA and OrfB, whereof OrfB displayed the highest conservation with values between 41 and 66 % homology on aa level (as determined by the program TFASTA based on Needleman-Wunsch algorithms). Considering that the transposase of IS3 elements is thought to be a transframe product of both orfA and orfB, the aa sequences of the putative fusion proteins were aligned and plotted in a dendrogramm using the HUSAR2.0 programme TREE (Feng and Doolittle, 1987). To summarize the phylogenetic relationship among the compared IS and to place IS1222, the dendrogramm is shown in Fig. 3.
The current model for ribosomal frameshifting implies that all factors decreasing the speed of the elongating ribosome along the mRNA, such as RBS, hungry codons and secondary structures, which have to be unwound, result in increased frameshifting at slippery sites (Polard et al., 1991; Sekine et al., 1994). In IS1222 a 5'-478AAAG481-3' motif was found, followed by a potential pseudoknot structure with a spacing of 6 nt, as marked in Fig. 2., displaying a high similarity to the site for frameshifting in IS3. In contrast to the heptanucleotide motif (A7, A6G respectively) of IS911 and IS150 (Polard et al., 1991; Vögele et al., 1991; Chandler and Fayet, 1993), the IS3 A4G homonucleotide sequence is combined with an mRNA secondary structure forming a pseudoknot. Located 5 nt downstream of the A4G motif this structure was shown to be necessary for ribosomal slippage on the mRNA of -1 in frame to orfB (Sekine et al., 1994). In the pseudoknot the 5' part of the second stem pairs within the loop region of the first stem-loop and borders with a spacing of 3 nt on the stem region of the first stem. Further increase in the efficieny of frameshifting is depending on the codon usage of individual host strains, when aminoacyl-tRNA limitation causes the ribosome to halt on the mRNA at a hungry codon and stimulates frameshifting (Kolor et al., 1992; Chandler and Fayet, 1993). At the postulated frameshift site IS1222 exhibits a 5'-479AAG481-3' codon that was shown to promote frameshifting in Escherichia coli - host strain of IS3. The lack of AAG-tRNA:Lys is compensated by a mismatched AAA-tRNA:Lys or causes ribosomal frameshifting as in the dnaX-gene (Tsuchihashi and Brown, 1992).
(e) Distribution of IS1222
The distribution and copy number were determined by hybridization with a DNA probe specific to IS1222. Physical mapping and further hybridization experiments under high stringent conditions revealed four copies of IS1222 in E. agglomerans 339, one located on the chromosome and three on the pEA9 plasmid. To determine the distribution of IS1222 among divergent bacterial strains, representatives for Enterobacter-related species and several unrelated taxonomic genera were tested for hybridization signals in dot and Southern blots (Table II). According to these early data, only Enterobacter strains closely related to E. agglomerans 339 showed copies of IS1222.
Considering the divergent evolution of IS by the adaption to individual host systems resulting in changes on DNA level (Lawrence et al., 1992), a detailed analysis of IS and their distribution can be used as a means for the classification of bacterial strains, taking into account (i) the presence and copy-number in a given strain and (ii) the homology on nt level among isoforms of an IS. This method was already applied to classify and identify Shigella and Escherichia coli strains isolated from various sources (Matsutani et al., 1993). According to the data obtained for IS1222, indicating a limited distribution among certain Enterobacter strains, this IS might also prove to be a tool for taxonomic classification of host strains among Enterobacteriaceae.
(f) Conclusions
(1) According to sequence alignments and similar structural features such as terminal IR, IS1222 belonged to a subclass within the IS3 family of bacterial IS, together with IS407, ISR1 and IS476 (Fiandt et al., 1972; Prère et al., 1990; Wood et al., 1991).
(2) Similar as demonstrated for IS3 (Sekine et al., 1994), the putative site for ribosomal frameshifting in IS1222 consisted of a slippery codon (homonucleotide motif) followed by a pseudoknot structure.
(3) The distribution of IS1222 appeared to be limited to a narrow host range among closely related Enterobacter strains.
Annotation: As suggested by Berge et al. (1991) and based upon recent studies on physiological and molecular level, E. agglomerans 339, a host strain of IS1222, will be reclassified as Rahnella aquatilis (S. Selenska-Pobell, personal communication).
We thank Prof. Dr. W. Klingmüller for giving us the possibility to perform this study at the Department for Genetics, University of Bayreuth, for his support and creative discussions. The pTGL274 plasmid containing IS407 was kindly provided by Dr. T. G. Lessie, Department of Microbiology, University of Massachusetts. We also thank our colleagues A. Lechler, G. Romanowski, S. Selenska-Pobell and U. Zink for their professional aid and helpful discussions. This work was supported by a scholarship for F.-M. L. from the Richard-Winter-Foundation.
Berge, O., Heulin, T., Achouak, W., Richard C., Bally R. and Balandreau, J.: Rahnella aquatilis, a nitrogen-fixing bacterium associated with the rhizosphere of wheat and maize. Can. J. Microbiol. 37 (1991) 195-203.
Cannon, F.C., Riedel, G.E. and Ausubel, F.M.: Overlapping segments of Klebsiella pneumoniae nif DNA cloned and characterized. Mol. Gen. Genet. 174 (1979) 59-66.
Chandler, M. and Fayet, O.: Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 7 (1993) 497-503.
Devereux, J., Haeberli, P. and Smithies, O.: A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12 (1984) 387-395.
Dodd, I.B. and Egan, J.B.: Systematic method for the detection of potential l-Cro-like DNA-binding regions in proteins. J. Mol. Biol. 194 (1987) 557-569.
Fayet, O., Ramond, P., Polard, P., Prère, M.F. and Chandler, M.: Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol. Microbiol. 4 (1990) 1771-1777.
Feng, D.F. and Doolittle, R.F.: Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evolution 25 (1987) 351-360.
Fiandt, M., Szybalski, W. and Malamy, M.H.: Polar mutations in lac, gal and Phage l consist of a few IS-DNA sequences inserted with either orientation. Mol. Gen. Genet. 119 (1972) 201-222.
Huber, I. and Selenska-Pobell, S.: Pulsed field gel electrophoresis fingerprinting, rrn loci number and genome size estimation of several Rhizobium galegae strains. J. Appl. Bacteriol. (1994, in press).
Khan, E., Mack, J.P.G., Katz, R.A., Kulkowsky, J. and Skalka, A.M.: Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 19 (1991) 851-860.
Kleeberger, A., Castorph, H. and Klingmüller, W.: The rhizosphere microflora of wheat and barley with special reference to gram-negative bacteria. Arch. Microbiol. 136 (1983) 306-311.
Klingmüller, W., Herterich, S., and Min, B.W.: Selftransmissible nif plasmids in Enterobacter. In: Skinner, F.A. and Uomala, P. (Eds.), Nitrogen Fixation with Non-Legumes. Kluwer Academic Publishers, 1989, pp. 173-178.
Kolor, K., Lindsley, D., and Gallant J.A.: On the role of the P-site in leftward ribosome frameshifting at a hungry codon. J. Mol. Biol. 230 (1993) 1-5.
Lawrence, J.G., Ochman, H. and Hartl, D.L.: The evolution of insertion sequences within enteric bacteria. Genetics 131 (1992) 9-20.
Matsutani, S. and Ohtsubo, E.:. Distribution of the Shigella sonnei insertion elements in Enterobacteriaceae. Gene 127 (1993) 111-115.
Polard, P., Prère, M.F., Chandler, M. and Fayet, O.: Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol. 222 (1991) 465-477.
Prère, M., Chandler, M. and Fayet, O.: Transposition in Shigella dysenteriae: Isolation and analysis of IS911, a member of the IS3 group of insertion sequences. J. Bacteriol. 172 (1990) 4090-4099.
Ruppel, S., Hecht-Buchholz, C., Remus, R., Ortmann, U. and Schmelzer, R.: Settlement of the diazotrophic, phytoeffective bacterial strain Pantoea agglomerans on and within winter wheat: an investigation using ELISA and transmission electron microscopy. Plant and Soil 145 (1992) 261-273.
Schwartz, E., Kröger, M. and Rak, B.: IS150: Distribution, nucleotide sequence and phylogenetic relationship of a new E. coli insertion element. Nucleic Acids Res. 16 (1988) 6789-6802.
Sekine, Y., Eisaki, N. and Ohtsubo, E.: Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235 (1994) 1406-1420.
Singh, M., Kleeberger, A. and Klingmüller, W.: Location of nitrogen fixation (nif) genes on indigenous plasmids of Enterobacter agglomerans. Mol. Gen. Genet. 190 (1983) 373-378.
Steibl, H.-D.: Ph.D.-thesis. University of Bayreuth, D-95440 Bayreuth, Germany (in preparation) 1994.
Steibl, H.-D., Siddavattam, D. and Klingmüller, W.: Similar nif-clusters on dissimilar plasmids of Enterobacter agglomerans strains. In: Palacios, R., Mora, J. and Newton, W. (Eds.), New Horizons in Nitrogen Fixation. Kluwer Academic Publishers, 1993, p. 652.
Tsuchihashi, Z. and Brown, P.O.: Sequence requirements for efficient translational frameshifting in the E. coli dnaX gene and the role of an unstable interaction between tRNA: Lys and an AAG Lysine codon. Genes Dev. 6 (1992) 511-519.
Vögele, K., Schwartz, E., Welz, C., Schiltz, E. and Rak B.: High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 19 (1991) 4377-4385.
Wood, M.S., Byrne, A. and Lessie, T.G.: IS406 and IS407, two gene-activating insertion sequences from Pseudomonas cepacia. Gene 105 (1991) 101-105.
Zuker, M. and Stiegler, P.: Optimal computer folding of large RNA sequences using thermodynamics and auxiliary Information. Nucleic Acids Res. 9 (1981) 133-148.
Steibl and Lewecke, Fig. 1.
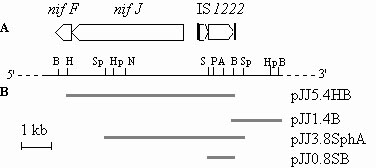
Fig. 1. Map and cloning-strategy for IS1222. (A) Physical map and organization of the pEA9 nif 3'-flanking region showing the location of IS1222 relative to nifF and nifJ. Restriction endonucleases are abbreviated as follows: A, ApaI; B, BamHI; H, HindIII; Hp, HpaI; N, NotI; P, PstI; S, SalI; Sp, SphI. (B) Size and position of cloned restriction fragments in recombinant plasmids. The 5.4-kb HindIII-BamHI fragment was subcloned into the sequencing vector pUC18 generating the recombinant plasmid pJJ5.4HB. Sequence analysis showed the BamHI restriction site close to the b terminus of the postulated IS element. To obtain the complete nt sequence of IS1222, the 3'-adjacent 1.4-kb BamHI fragment from pHD54 was also cloned into pUC18, the construct named pJJ1.4B. Plasmid pJJ5.4HB was linearized at restriction sites close to the 3' end of the cloned fragment, treated with exonucleaseIII and S1 nuclease to create unidirectional, nested deletions. Plasmid DNA from pJJ1.4B, pJJ5.4HB and its deletion-derivatives was sequenced according to the chain-termination procedure using the M13 universal and reverse primers. For obtaining a plasmid containing the entire IS1222 the 3.8-kb SphI fragment from pHD54 was also cloned into pUC18. To sequence the complementary strand and to ease the later preparation of IS1222-specific probe DNA the 0.8-kb SalI-BamHI fragment from pJJ5.4HB containing exclusively parts of the IS was subcloned into the multicopy vector pUC18 as well.
Steibl and Lewecke, Fig. 2.

Fig. 2. Nucleotide sequence of IS1222 and flanking pEA9 DNA. The IS1222 sequence is printed for both strands; the flanking regions as 5'® 3' strand only. The inverse repetitive termini are printed in boldface letters, marked with IRL and IRR. Internal IR are indicated by arrows above; IR forming a potential pseudoknot are marked 1a, 1b, 2a, 2b and the A3G frameshift window by a double line. Sequences homologous to the s70 promoter consensus are underlined (-35, -10). Start and stop codons of orfA and orfB are typed in boldface letters. Both ORFs were preceded by RBS. The translation of orfA and orfB is shown below the nt sequence. IS1222 is listed in the GenBank library under accession No. X78052.
Steibl and Lewecke, Fig. 3.
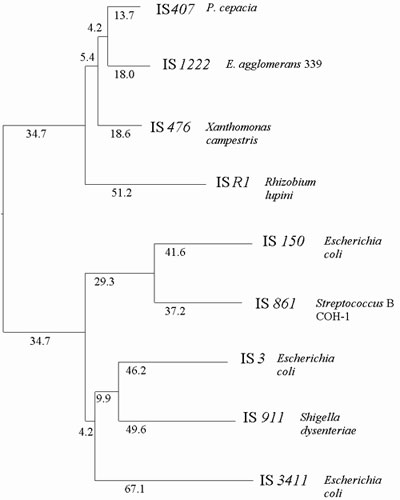
Fig. 3. Phylogenetic relationship of selected IS3 elements based on a multiple alignment of the translated, transframe orfA-orfB aa sequences using the HUSAR program TREE (Feng and Doolittle, 1987). Small numbers beside the branches represent values of dissimilarity as calculated from the alignment data matrix. Together with IS407, IS476 and ISR1, IS1222 forms a closely related group among IS3 elements. This grouping, reflecting the evolution of IS, does not correspond to the phylogeny of the host organisms indicating the influence of horizontal transfer of IS (note: host organisms of the listed IS range among a variety of different taxonomic genera).
Steibl and Lewecke
TABLE I
Conservation of terminal IR among IS3 elementsa
___________________________________________________________________________
IS IR Sequence of terminal IR
___________________________________________________________________________
|
IS150 |
R |
TGTACTGACCCCAAAAAGTTGGACAGTTAAACACGA |
||||||||||
|
L |
TGTACTGCACCCATTTTGTTGGACGATGAAATGGAA |
|||||||||||
|
IS861 |
R |
TGTACTGACCCCCAAAAGTTGGACAATTTATTTTAAG |
||||||||||
|
L |
TGAACTGCACCCCAAAAGTTAGACAAAAAATTTAACG |
|||||||||||
|
IS3411 |
R |
TGAACCGCCCCGGGTTTCCTGGAGAGTGTTTTATCTGTGA |
||||||||||
|
L |
TGAACCGCCCCGGGAATCCTGGAGACTAAACTTCCTGAGA |
|||||||||||
|
IS911 |
R |
TGAAGTGGCACACTGAATTTGGCCACCTGAACAGAGGT |
||||||||||
|
L |
TGAAGTGGTCAACAAAAACTGGCCACCGAGTTAGAGTT |
|||||||||||
|
IS3 |
R |
TGATCCTACCCACGTAATATGGACACAGGCCTAAGCGAG |
||||||||||
|
L |
TGATCTTACCCAGCAATAGTGGACACGCGGCTAAGTGAG |
|||||||||||
|
IS476 |
R |
TGACCTGCCCCCATCGT CCGTACCAGCTGAAGCTATAAA |
||||||||||
|
L |
TGACCTGCCCCCACTGAGCCGTACCAGTGATTACTGATAA |
|||||||||||
|
IS407 |
R |
TGACCTGCCCCCATCAATAGGGCCAATG GGC TCTAGCAAAGTCC |
||||||||||
|
L |
TGACCTGCCCCCTGCAAACAGGGCCAGCCGGAGTCTAGTAAAGTTC |
|||||||||||
|
ISR1 |
R |
TGACGTGACCCCCGTTT CTGATCCAGCCATAACGAGAGTCC |
||||||||||
|
L |
TGACGTGACCCCCTGAAACTCCTCCAGGAATAGCTAGAGTCC |
|||||||||||
|
IS1222 |
R |
TGACCTGCCCCCAG GATTAGATACAACGCTCACTTAGTAATGTC |
||||||||||
|
L |
TGACCTGCTCCCAGTTGATTAG TACACCCCGATGTTAGTAATGTC |
|||||||||||
|
Consensus |
TGA CTG CCCC TGG C |
|||||||||||
___________________________________________________________________________
a
References and emendation of nt sequences for the listed IS in Prère et al. (1990).R and L define the right and left IR. Mismatches between the left and right IR are printed in boldface letters. Highly conserved nt of the IR are given in the consensus showing a high conservation of the termini. Note the 100% conservation of the 5'-TG-3' terminus. This terminal dinucleotide was shown to be the substrate for the OrfB and retroviral integrase, carrying the D(35)E sequence motif (Kahn et al., 1990).
Steibl and Lewecke
TABLE II
Strains tested for the presence of IS1222
___________________________________________________________________________
Hybridization to IS-specific probe:
(A) positive (B) negative
___________________________________________________________________________
|
strain |
reference |
strain |
reference |
|
E. cloacae |
A. Glatzlea |
Azospirillum brasilense |
DSM 1843c |
|
E. agglomerans 19-78 |
Berge et al. (1991) |
Azotobacter vinelandii |
DSM 85 |
|
E. agglomerans 243 |
Kleeberger et al. (1983) |
Bacillus subtilis 168 |
DSM 347 |
|
E. agglomerans 335 |
Kleeberger et al. (1983) |
Enterobacter aerogenes |
d |
|
E. agglomerans 339 |
Kleeberger et al. (1983) |
E. agglomerans 333 |
Kleeberger et al. (1983) |
|
R. aquatilis 2-87 |
Berge et al. (1991) |
E. agglomerans 334 |
Kleeberger et al. (1983) |
|
R. aquatilis 3-88 |
Berge et al. (1991) |
Escherichia coli B |
d |
|
R. aquatilis |
ATCC 33071b |
Klebsiella pneumoniae M5aI |
Cannon et al. (1979) |
|
Pantoea agglomerans |
Ruppel et al. (1992) |
Paracoccus denitrificans |
DSM 65 |
|
Proteus vulgaris |
DSM 30118 |
||
|
Pseudomonas aeruginosa PAO5 |
W. Schilfe |
||
|
R. aquatilis |
ATCC 33989 |
||
|
Rhizobium galegae |
Huber et al. (1994) |
___________________________________________________________________________
a University of Stuttgart, Stuttgart/Hohenheim, Germany; b ATCC, American Type Culture Collection, Rockville, MD, USA; c DSM, Deutsche Sammlung für Mikroorganismen, Göttingen, Germany; d Department for Genetics, University of Bayreuth, Germany; and e TFH Berlin, Germany. Rahnella is abbreviated as R..
Methods: Dot blots of total DNA from the strains listed above were hybridized and to obtain characteristic fingerprints for strains with a positve signal, total DNA was restricted by BamHI, separated on agarose gels, vacuum-blotted onto Nylon membranes and hybridized with the IS1222-specific DNA probe according to Southern. The probe was prepared by labelling the SalI-BamHI insert from pJJ0.8SB with digoxigenin. To obtain IS1222-specific signals, hybridization and washing was performed under conditions of high stringency. No crosshybridization with related IS was detected using the closely related IS407 as negative reference (plasmid pTGL274; Wood et al., 1991). For the detection of probe/target hybrids the DNA Labeling and Detection Kit nonradioactive (Boehringer, Mannheim, Germany) was used. Note that all strains hybridizing belong to the taxonomic group of Enterobacteriaceae.
<< nach oben
zurück